I am Haoyang Liu, a PhD student in Informatics at the School of Information Sciences, University of Illinois Urbana-Champaign. I am fortunate to be advised by Prof. Bertram Ludäscher. Prior to that, I had the opportunity to work with Prof. Haohan Wang on trustworthy ML, Prof. Halil Kilicoglu on biomedical NLP, and interned at BNRist, Tsinghua University advised by Prof. Chunxiao Xing.
My research aims to develop trustworthy machine learning methods and systems. I have studied ways to evaluate and improve model robustness (ICLR 2024, CPAL 2025 Oral), dataset distillation for more compute- and data-efficient learning (ICCV 2025) that also improves robustness (AAAI 2025), and LLM agents for robust automation of biomedical data analysis workflows (MLCB 2025 Oral). Currently, I am interested in combining formal methods with LLM agents for trustworthy reasoning and automation, with applications in biomedical and legal domains.
Selected Publications
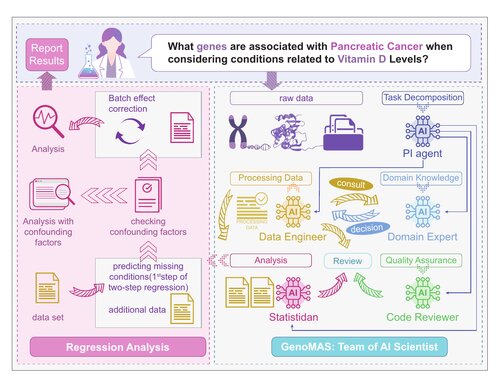
GenoMAS: A Multi-Agent Framework for Scientific Discovery via Code-Driven Gene Expression Analysis
Haoyang Liu, Yijiang Li, Haohan Wang
arXiv preprint, 2025
A multi-agent framework that combines workflow controllability with agentic autonomy for automated gene expression analysis. Features a guided-planning mechanism that enables agents to dynamically advance, revise, or backtrack while maintaining logical coherence.
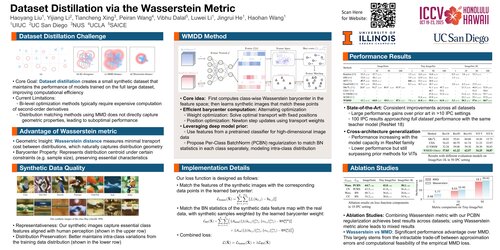
Dataset Distillation via the Wasserstein Metric
Haoyang Liu, Yijiang Li, Tiancheng Xing, Peiran Wang, Vibhu Dalal, Luwei Li, Jingrui He, Haohan Wang
IEEE/CVF International Conference on Computer Vision (ICCV), 2025
Leverages optimal transport theory for dataset distillation, using Wasserstein barycenter to preserve geometric characteristics of the original data distribution. Achieves consistent state-of-the-art performance across multiple high-resolution benchmarks.
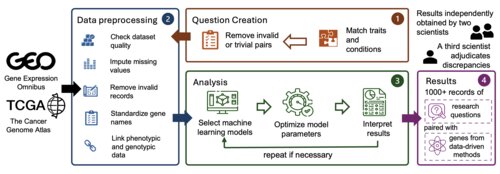
GenoTEX: An LLM Agent Benchmark for Automated Gene Expression Data Analysis ⭐ Oral (14.4%)
Haoyang Liu, Shuyu Chen, Ye Zhang, Haohan Wang
The 20th Machine Learning in Computational Biology Conference (MLCB), 2025
A benchmark for evaluating LLM agents on automated gene expression analysis, featuring 913 real-world datasets with realistic complexities covering 132 human traits.
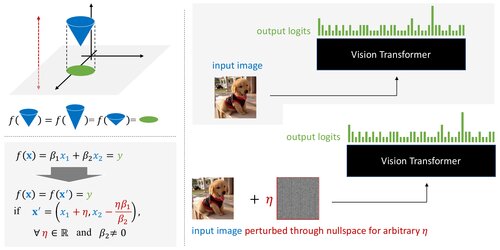
Approximate Nullspace Augmented Finetuning for Robust Vision Transformers ⭐ Oral (12.7%)
Haoyang Liu, Aditya Singh, Yijiang Li, Haohan Wang
The 2nd Conference on Parsimony and Learning (CPAL), 2025
Generalizes the nullspace concept from linear layers to non-linear ViT encoder blocks, enhancing both adversarial and natural robustness while maintaining clean accuracy.
Towards Adversarially Robust Dataset Distillation by Curvature Regularization
Eric Xue, Yijiang Li, Haoyang Liu, Peiran Wang, Yifan Shen, Haohan Wang
The 39th AAAI Conference on Artificial Intelligence (AAAI), 2025
Embeds adversarial robustness into distilled datasets via curvature regularization. Minimizes loss landscape curvature during distillation, reducing adversarial vulnerability with significantly less overhead than adversarial training.
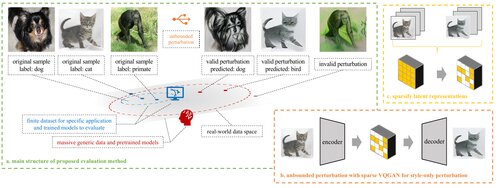
Foundation Model-oriented Robustness: Robust Image Model Evaluation with Pretrained Models
Peiyan Zhang, Haoyang Liu, Chaozhuo Li, Xing Xie, Sunghun Kim, Haohan Wang
The 12th International Conference on Learning Representations (ICLR), 2024
A dynamic robustness evaluation protocol using foundation models as surrogate oracles, enabling evaluation beyond fixed benchmarks through semantically bounded perturbations.
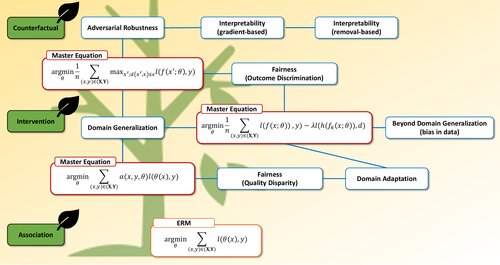
Towards Trustworthy and Aligned Machine Learning: A Data-centric Survey with Causality Perspectives
Haoyang Liu, Maheep Chaudhary, Haohan Wang
arXiv preprint, 2023
A data-centric survey on trustworthy ML covering robustness, security, interpretability, and fairness, unified through Pearl’s causal hierarchy.
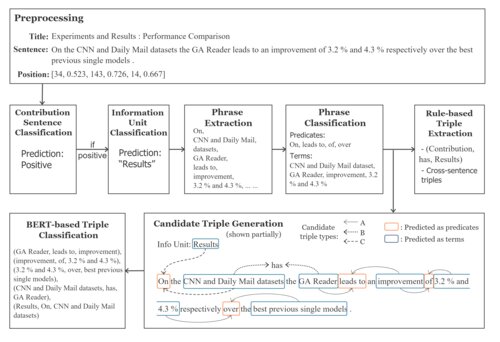
UIUC_BioNLP at SemEval-2021 Task 11: A Cascade of Neural Models for Structuring Scholarly NLP Contributions 🏆 Best System Paper (1/175)
Haoyang Liu, Janina Sarol, Halil Kilicoglu
The 15th International Workshop on Semantic Evaluation (SemEval), 2021
A neuro-symbolic approach combining BERT-based classifiers with rule-based systems for extracting structured contributions from NLP papers. Ranked first overall across all evaluation phases.
Honors and Awards
- 2022, List of Teachers Ranked as Excellent, University of Illinois Urbana-Champaign
- 2021, Best System Paper Award (1st place out of 175 submissions), SemEval 2021
Education
- 2020.06 - Present, Ph.D. in Informatics, University of Illinois Urbana-Champaign, USA
- 2020.01 - 2020.05, Visiting Student in Computer Science, Illinois Institute of Technology, USA
- 2016.09 - 2020.06, B.S. in Telecommunication Engineering, Beijing University of Posts and Telecommunications, China
Invited Talks
- 2025.03, Approximate Nullspace Augmented Finetuning for Robust Vision Transformers, CPAL 2025 Highlight Talk, Stanford University
- 2021.11, Information Extraction from NLP Literature, Research Showcase, UIUC School of Information Sciences
- 2021.08, UIUC_BioNLP System at SemEval-2021, 15th International Workshop on Semantic Evaluation (Virtual)
Teaching and Services
Teaching
- FA22 - SP23, Instructor of Record, IS 203: Analytical Foundations for Information Problems, UIUC
- SP24, Teaching Assistant, IS 597 TML: Trustworthy Machine Learning, UIUC
- FA23, Teaching Assistant, IS 507: Data, Statistical Models, and Information, UIUC
Academic Services
- Conference Reviewer: ICML (2023-2024), NeurIPS (2024), ICLR (2024-2026), CVPR (2025-2026)
- Program Committee Member: AAAI 2026, KDD 2026 D&B Track
Internships
- 2019.03 - 2019.09, Research Intern, Beijing National Research Center for Information Science and Technology (BNRist), Tsinghua University, Beijing, China
- Advisor: Prof. Chunxiao Xing
- Developed automated system to construct medical knowledge graphs from electronic health records
- Built multimodal neural network for diabetes risk prediction from clinical notes and tabular data